2014 DURF CONFERENCE POSTER
-
Upload
sudeep-pisipaty -
Category
Documents
-
view
26 -
download
0
Transcript of 2014 DURF CONFERENCE POSTER

Protein function dictates cellular function and is a cornerstone of learning and memory in the brain. A major regulator of neural protein synthesis is mammalian target of rapamycin (mTOR) complex 1 (mTORC1). Studies have shown that mTORC1 is dysregulated in many forms of autism, including Fragile X Syndrome (FXS) and Tuberous Sclerosis Complex (TSC). An enzyme located downstream of mTORC1 is p70-S6 ribosomal Kinase 1 (S6K1) which is involved in the modulation of several steps of protein synthesis and is deregulated in FXS condition, and perhaps other major forms of autism. Manipulation of S6K1 thereby bears therapeutic value not only in FXS but also in autism broadly. Small genetic variations (SNVs) in S6K1 have been uncovered in autism recently. The goal of this research project is to characterize one SNV, which we have found in autistic patients in S6K1. We mutagenized human S6K1 cDNA and introduced our SNV of interest and expressed both wild-type (WT) and mutant (EQ) cDNA in HEK 293 cells. Following insulin stimulation to activate mTORC1-driven translation we found that the mutant was refractory to act ivat ion as measured from the phosphorylation of: eEF2 and S6240. We are currently investigating the impact of this mutation on basal levels of translation in HEK 293 cells. We believe that further structure function analysis of this and other mutants of S6K1 prevalent in autistic individuals would be important to understand the variation of translation control in autism.
Role of p70 Ribosomal S6 Kinase 1 Mutations in Autism Sudeep Pisipaty1, Aditi Bhattacharya1, Prakash Subramanyam2, Henry Colecraft2, Ivan Iossifov3 and Eric Klann1
1Center for Neural Science, New York University, NY, NY, USA
2Dept. of Physiology and Cellular Biophysics, Columbia University Medical Center, New York, NY 3Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Abstract
Acknowledgments
This work has been made possible by the New York University Dean’s Undergraduate Research Grant, issued for the Fall of 2013. AB is a recipient of the Charles H. Revson Fellowship.
Conclusions and Future Directions
Mammalian Target of Rapamycin (mTOR) Pathway
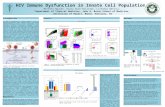
The figure outlines the mTOR pathway, which is critical in cellular functions that govern and control cell growth, proliferation survival and protein synthesis.
Single Nucleotide Variations in Human S6K1
Domain composition and SNVs in human p70 ribosomal S6K1 (RPS6KB1). A) Domains of human S6K1. Note that kinase domain is not drawn to scale (adapted from Magnuson et al., 2012). B) Distribution of SNVs as reported by NIH SNP database and those found in the Simon Simplex Collection (SSC), screened in collaboration with Dr. Iossifov. The missense variation in the SSC is highlighted in red.
Basal levels of phophorylation of S6K1 downstream targets is effected by the E44Q mutation
imm
unor
eact
ivity
(r
elat
ive
to W
T t=
0min
s)
time (min)
*
WT EQ
peEF2
eEF2
0 20 40 60 0 20 40 60
imm
unor
eact
ivity
(r
elat
ive
to W
T t=
0min
s)
WT EQ 0 20 40 60 0 20 40 60
pS6 240-44
S6
* *
time (min)
A
B
Figure A- Time course of phosphorylation of eEF2 is shown across cells that have been transfected with wild-type and mutant S6K1 constructs. N=4 lysates prepared from cells of different passages. * denotes p<0.05 by two-tailed t-test. Figure B- Time course of phosphorylation of S6 (240-44) is shown across cells that have been transfected with wild-type and mutant S6K1 constructs. N=4 lysates prepared from cells of different passages. * denotes p<0.05 by two-tailed t-test.
*
S6K1 E-Q
S6K1 WT
aver
age
fluor
esce
nce
(a.u
)
No effect seen in basal protein synthesis between wild-type and E44Q transfected
cells.
HEK 293 cells expressing both E-Q and wild-type cells were tagged with non-canonical amino acids (FUNCAT method, Dieterich et al, 2010). This tagged newly synthesized proteins (red fluorescence). An additional green-fluorescent is shown in the green channel expressing EGFP. White markers are placed denoting same cells in both the green and the red channels. N=22 cells quantified in one experiment.
• Database known as Simon Simplex Collection, containing DNA information of 2700 families was analyzed for single nucleotide variations (SNVs) for novel genetic variations only in Autistic child
• A missense mutation, E44Q, was identified in S6K1 • Human S6K1 cDNA was engineered to contain a c-terminal
EGFP fusion and the E44Q mutation was introduced this construct.
• In our studies of eEF2 and S6, our mutants show lower levels of activity than the wild-type.
• When we checked for differences in basal protein synthesis using FUNCAT, we found no appreciable differences between wild-type and EQ.
• Standardize our FUNCAT experiment to be able to detect
discernable differences between our wild-type and EQ mutant.
• Examine the effect of the EQ mutation on S6K1 phosphorylation.
• Express the EQ mutants and wild-type constructs in neurons and examine effect on phosphorylation of eEF2 and S6, protein synthesis, and any other phenotypes.
Insulin
Insulin
Method
S6K1 Gene S6K1 Mutant
Transfection
HEK-293 Cells
Creation of EGFP fusion construct and mutagenesis to generate EQ
construct
E44Q